Research Interests
I’m interested in how changes to DNA and chromatin structure unfold across multiple biological scales — from molecular mechanisms in individual cells, to long-term effects on development and evolutionary outcomes across species.
Molecular Dynamics During Cellular Processes
What is it like inside the nucleus of a cell? How does DNA behave at the molecular level — how does it move, coil, and interact within the dense, dynamic space of chromatin? I’m drawn to questions about the physical nature of chromosomes and the balance between randomness and regulation in the genome’s internal mechanics.
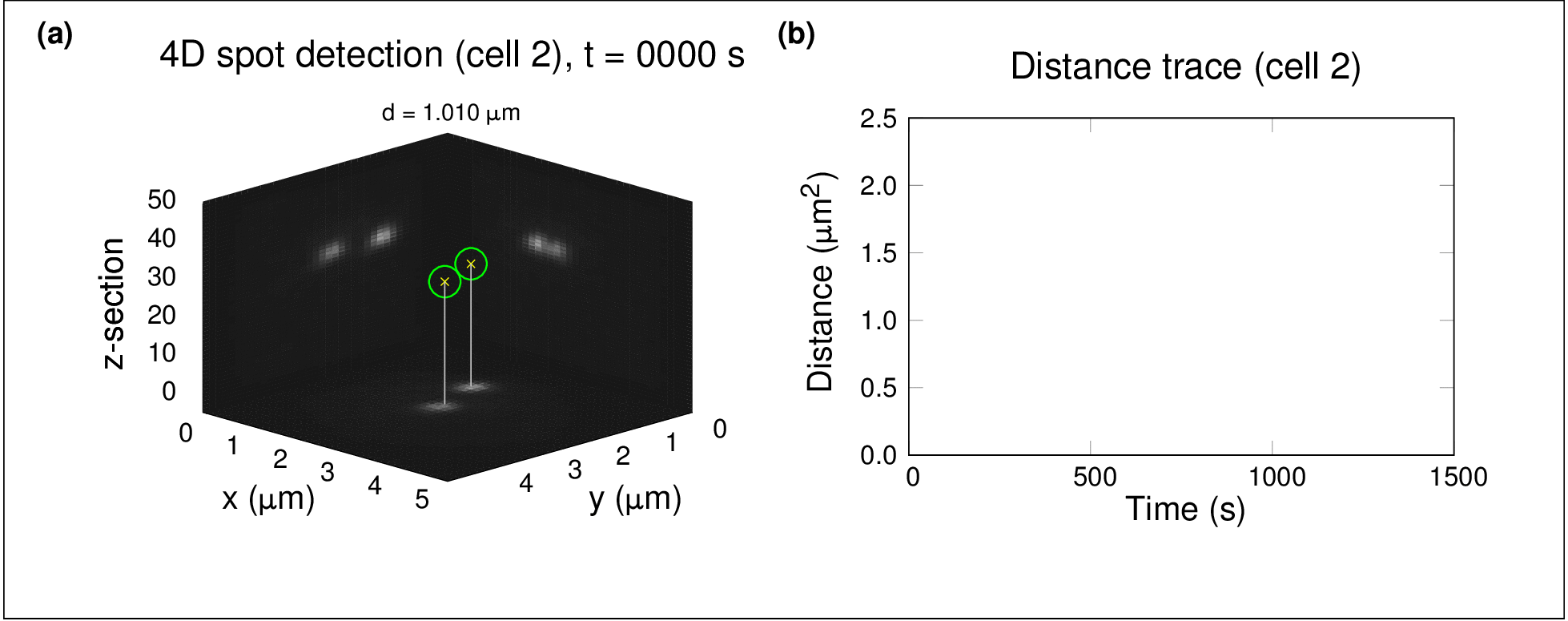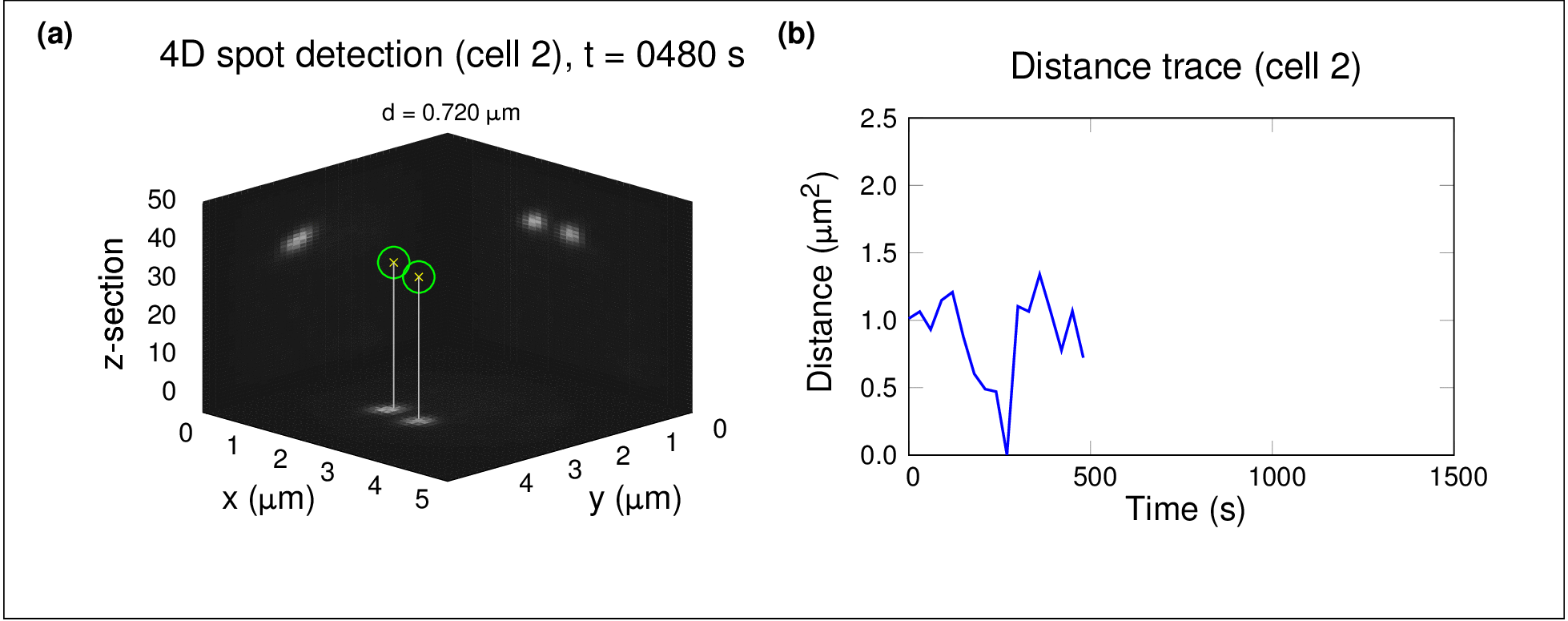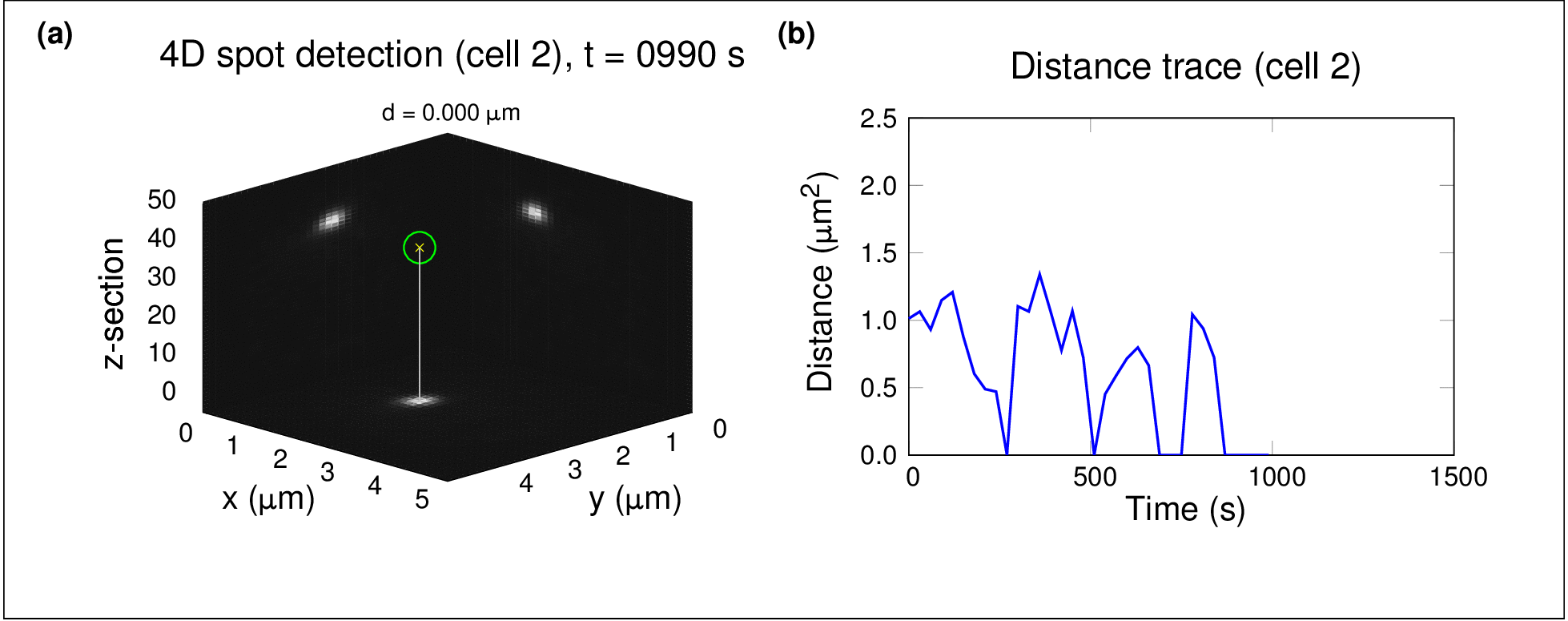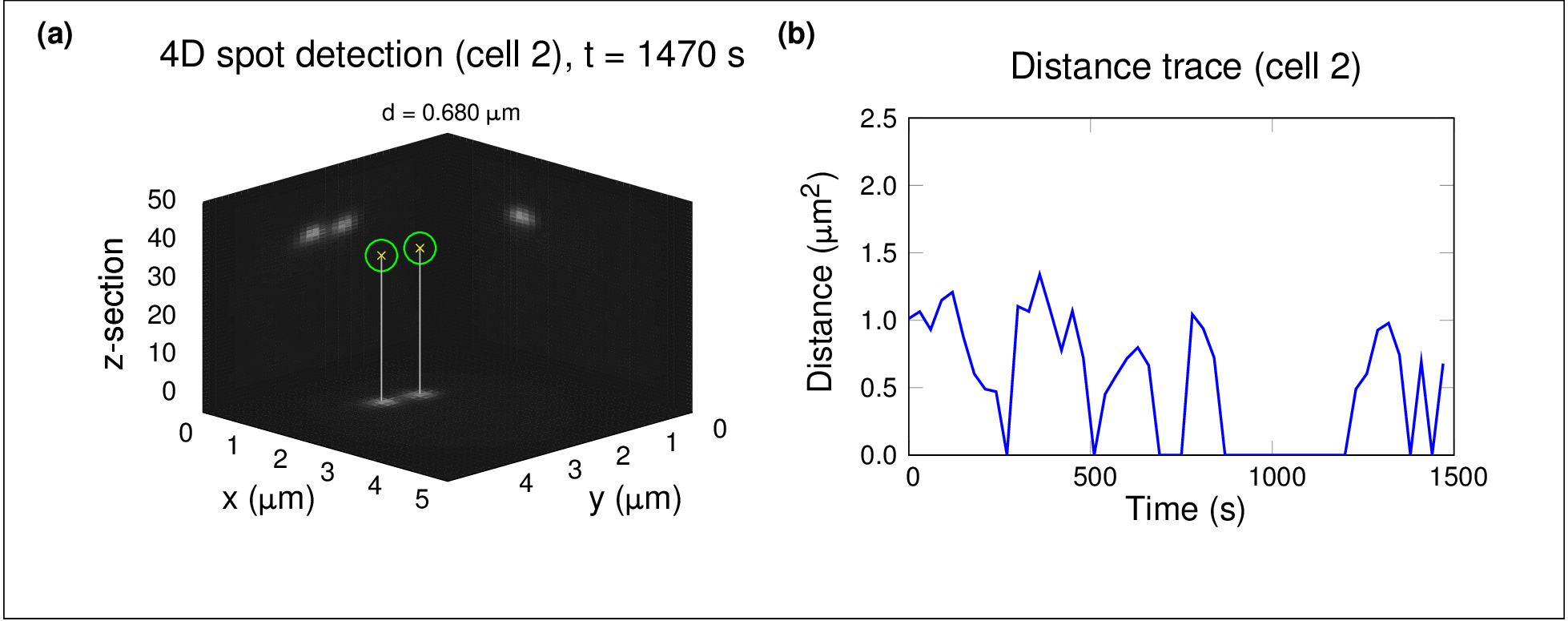
In one project, I used high-throughput spinning-disc confocal imaging and single-particle tracking to study chromosome dynamics during meiosis in yeast. By analysing fluorescently labelled chromosomes over time, we showed that diffusion alone was sufficient for homologous chromosomes to find and pair with each other — no active force generation was necessary. This raised fascinating questions about the role of stochastic processes in organising biological systems.
Environmental Effects on Development
How do organisms respond to their environment — not just in immediate ways, but in terms of development, reproduction, and long-term health? I’m especially interested in how early-life exposures shape phenotypes across the lifespan, and whether those influences can be passed on to the next generation.

In zebrafish, I’ve explored how environmental stress during embryogenesis — such as transient oxidative stress — leads to lasting reductions in adult fertility. I’ve also studied the impact of parental diet on gamete quality and offspring growth. These experiments ask to what extent today’s conditions influence not only our health, but also that of future generations.
- Newman, T., Jhinku, N., Meier, M., & Horsfield, J. (2016). Dietary intake influences adult fertility and offspring fitness in zebrafish. PLoS One, 11(11), e0166394.
- Newman, T., Carleton, C. R., Leeke, B., Hampton, M. B., & Horsfield, J. A. (2015). Embryonic oxidative stress results in reproductive impairment for adult zebrafish. Redox Biology, 6, 648-655.
Epigenetic Evolution in Mammalian Development
How do regulatory systems evolve? Where do new genes come from, and how does molecular complexity arise? I’m especially intrigued by epigenetic layers of control — like genomic imprinting — that shape gene expression in lineage-specific ways. Can we trace the history of mammalian traits through their epigenetic blueprints?
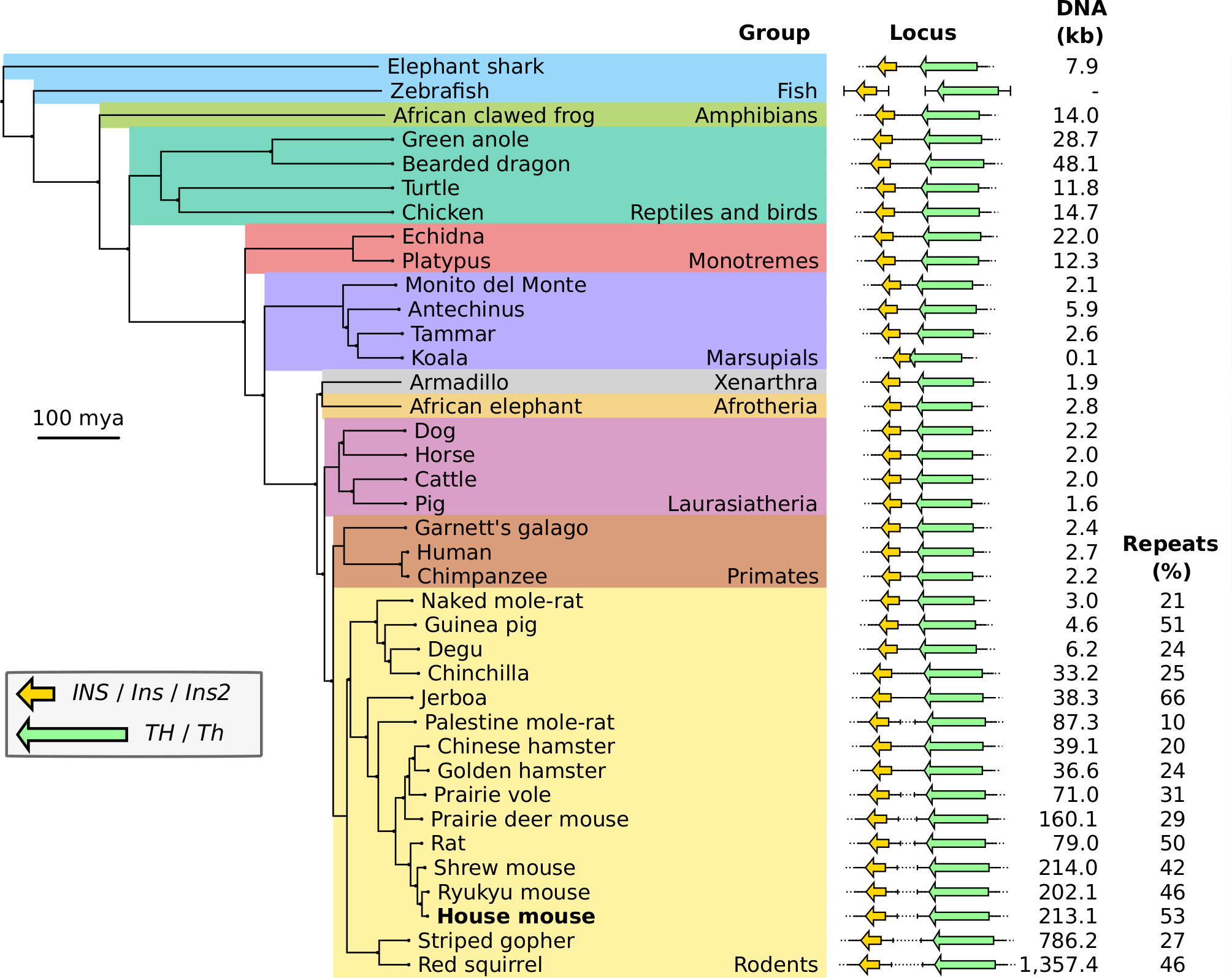
My research on imprinting in marsupials, especially wallabies, combines comparative genomics, DNA methylation, transcriptomics, and 3D genome structure. I’ve discovered new imprinted genes and identified lineage-specific imprinting events, offering insight into the evolutionary dynamics of epigenetic control. It’s remarkable how differently maternal and paternal alleles can behave — and how those differences contribute to our biology as mammals.
- Newman, T., Ishihara, T., Rizzoli, P., Xiao, L., Gouil, Q., Bond, D.M., Hore, T.A., Shaw, G., & Renfree, M.B. (2024). The PRKACB gene is imprinted in marsupials. Epigenetics & Chromatin, 17(1), 29.
- Newman, T., Ishihara, T., Shaw, G., & Renfree M.B. (2024). The structure of the TH/INS locus and the parental allele expressed are not conserved between mammals. Heredity, 133, 21-32.